Path Analysis
lecture_path.RmdEstimation
- Estimation is the math that goes on behind the scenes to give you parameter numbers
- Common types:
- Maximum Likelihood (ML)
- Asymptotically Distribution Free (ADF)
- Unweighted Least Squares (ULS)
- Two stage least squares (TSLS)
Maximum Likelihood
- Estimates are the ones that maximize the likelihood that the data were drawn from the population
- Normal theory method - multivariate normality is assumed to use ML
- Check your normality assumption!
- Other types of estimations may work better for non-normal endogenous variables
- Full information method - estimates are calculated all at the same
time
- Partial information methods calculate part of the estimates, then use those to calculate the rest
Maximum Likelihood
- Fit function - the relationship between the sample covariances and estimated covariances
- We want our fit function to be:
- High if we are measuring how much they match (goodness of fit)
- Low if we are measuring how much they mismatch (residuals)
Maximum Likelihood
- ML is an iterative process
- The computer calculates a possible start solution
- Then runs several times to create the largest ML match
- Start values:
- Usually generated by the computer
- You can enter values if you are having problems converging to a solution
Maximum Likelihood
- Scale free/invariant
- Means that if you change the scale with a linear transform, the model is still the same
- Assumes unstandardized start variables
- Otherwise you would be standardizing standardized estimates
- Interpretation of Estimates
- Loadings/path coefficients - just like regression coefficients
- Covariance/correlation - how much two items vary together
- Error variances tell you how much variance is not accounted for by the model (so you want to be small)
- SMCs are the amount of variance accounted for in a endogenous variable
Other Methods
- For continuous variables with normal distributions:
- Generalized Least Squares (GLS)
- Unweighted Least Squares (ULS)
- Fully Weighted Least Squares (WLS)
Generalized Least Squares
- Pros:
- Scale free
- Faster computation time
- Cons:
- Not commonly used? If this runs usually so does ML
Unweighted Least Squares
- Pros:
- Does not require positive definite matrices
- Robust initial estimates
- Cons:
- Not scale free
- Not as efficient
- All variables in the same scale
Other Methods
- Nonnormal data
- In ML, estimates might be accurate but standard errors may be large
- Model fit tends to be overestimated
- Corrected normal method - uses ML but then adjusts the SEs to be
normal (robust SE).
- Satorra-Bentler statistic: Adjusts the chi square value from standard ML by the degree of kurtosis/skew
- Corrected model test statistic
Asymptotically Distribution Free
- Some books call this arbitrary distribution free
- Estimates the skew/kurtosis in the data to generate a model
- May not converge because of number of parameters to estimate
Errors
- Inadmissible solutions - you get numbers in your output but clearly parameters are
- Heywood cases
- Parameter estimates are illogical (huge)
- Negative variance estimates
- Just variances, covariances can be negative
- Correlation estimates over 1 (SMCs)
Why Errors Happen
- Specification error
- Nonidentification
- Outliers
- Small samples
- Two indicators per latent (more is always better)
- Bad start values (especially for errors)
- Very low or high correlations (empirical under identification)
Path Models
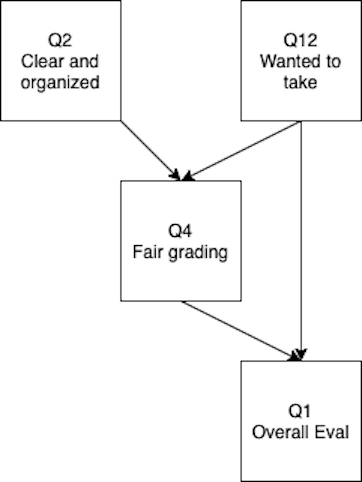
- Reminder:
- Circles are latent (unobserved) variables
- Squares are manifest (observed) variables
- Triangles can be used to represent intercepts (specific models use these)
- So what is a path model?
- A model with only manifest variables (measured/squares)
Path Models
- Straight arrows are “causal” or directional
- Non-standardized solution -> these are your b or slope values
- Standardized solution -> these are your beta values
- Curved arrows are non-directional
- Non-standardized -> covariance
- Standardized -> correlation
Path Models
- All endogenous variables have an error term
- All exogenous variables will have a variance
- All exogenous only variables are automatically correlated by lavaan
Solutions
- Why standardized?
- Allows you to compare path coefficients for strength of relationship
- Makes the non-directional relationships correlation (more interpretable)
- Matches the values you are used to thinking about (EFA)
- Why unstandardized?
- Allows you to estimate future scores and interpret path coefficients, in the original scale of the data
Let’s do it!
- Install
lavaan: latent variable analysis - Install
semPlot: helps us make pictures
install.packages("lavaan")
install.packages("semPlot")Syntax
- Note: there’s a cheat sheet guide on Canvas
- First, we will define a “model”
- Best practice: name the saved variable something like
name.modelwherenameis a descriptive name of the model you are building
- Best practice: name the saved variable something like
- Model syntax
-
~indicates a regression -
~~indicates a covariance/correlation -
=~indicates a latent variable
-
Syntax
- Automatically adds an error term to each endogenous variable
- Automatically constrains the path to 1 for you for the marker variable
- Automatically adds a covariance term between all exogenous variables
- Very useful for analyses we will get to later
- Not always required in path models, but you could constrain it to zero
- Important to note because it’s often unexpected
Estimate df
- Possible =
- Estimated:
- 4 regressions
- 2 error variances
- 2 variances
- 1 covariance
- DF = 10 - 9 = 1
Important Notes
eval.model
#> [1] "\nq4 ~ q12 + q2\nq1 ~ q4 + q12\n"- Note that
\nis code for new lines- These are important
- Spacing is not important
- You can use # in the code, it will ignore that line just like R
Run the Model
- We can use the
sem()function to apply the model to the data - Save the analysis, use good naming like
eval.output - The default estimator is normal theory maximum likelihood
- Use
?semto learn more about the options
eval.output <- sem(model = eval.model,
data = eval.data)View the Output
summary(eval.output)
#> lavaan 0.6-19 ended normally after 2 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 6
#>
#> Number of observations 3585
#>
#> Model Test User Model:
#>
#> Test statistic 2152.922
#> Degrees of freedom 1
#> P-value (Chi-square) 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|)
#> q4 ~
#> q12 0.096 0.008 12.479 0.000
#> q2 0.435 0.010 45.655 0.000
#> q1 ~
#> q4 0.840 0.017 49.923 0.000
#> q12 0.263 0.010 26.551 0.000
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|)
#> .q4 0.094 0.002 42.338 0.000
#> .q1 0.151 0.004 42.338 0.000Output
- Default output includes:
- Sample size
- Estimator
- Minimum function test statistic , df, p
- Unstandardized parameter estimates, standard error, Z, p
Improved Output
summary(eval.output,
standardized = TRUE, # for the standardized solution
fit.measures = TRUE, # for model fit
rsquare = TRUE) # for SMCs
#> lavaan 0.6-19 ended normally after 2 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 6
#>
#> Number of observations 3585
#>
#> Model Test User Model:
#>
#> Test statistic 2152.922
#> Degrees of freedom 1
#> P-value (Chi-square) 0.000
#>
#> Model Test Baseline Model:
#>
#> Test statistic 7293.347
#> Degrees of freedom 5
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 0.705
#> Tucker-Lewis Index (TLI) -0.476
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -2539.494
#> Loglikelihood unrestricted model (H1) -1463.033
#>
#> Akaike (AIC) 5090.988
#> Bayesian (BIC) 5128.095
#> Sample-size adjusted Bayesian (SABIC) 5109.030
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.775
#> 90 Percent confidence interval - lower 0.747
#> 90 Percent confidence interval - upper 0.802
#> P-value H_0: RMSEA <= 0.050 0.000
#> P-value H_0: RMSEA >= 0.080 1.000
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.105
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> q4 ~
#> q12 0.096 0.008 12.479 0.000 0.096 0.164
#> q2 0.435 0.010 45.655 0.000 0.435 0.598
#> q1 ~
#> q4 0.840 0.017 49.923 0.000 0.840 0.585
#> q12 0.263 0.010 26.551 0.000 0.263 0.311
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .q4 0.094 0.002 42.338 0.000 0.094 0.553
#> .q1 0.151 0.004 42.338 0.000 0.151 0.431
#>
#> R-Square:
#> Estimate
#> q4 0.447
#> q1 0.569Create a Picture
- Sometimes it’s hard to know if you have modeled what you intended to diagram
- Let’s create a plot to make sure it’s what we meant to do
library(semPlot)
semPaths(eval.output, # the analyzed model
whatLabels = "par", # what to add as the numbers, std for standardized
edge.label.cex = 1, # make the font bigger
layout = "spring") # change the layout tree, circle, spring, tree2, circle2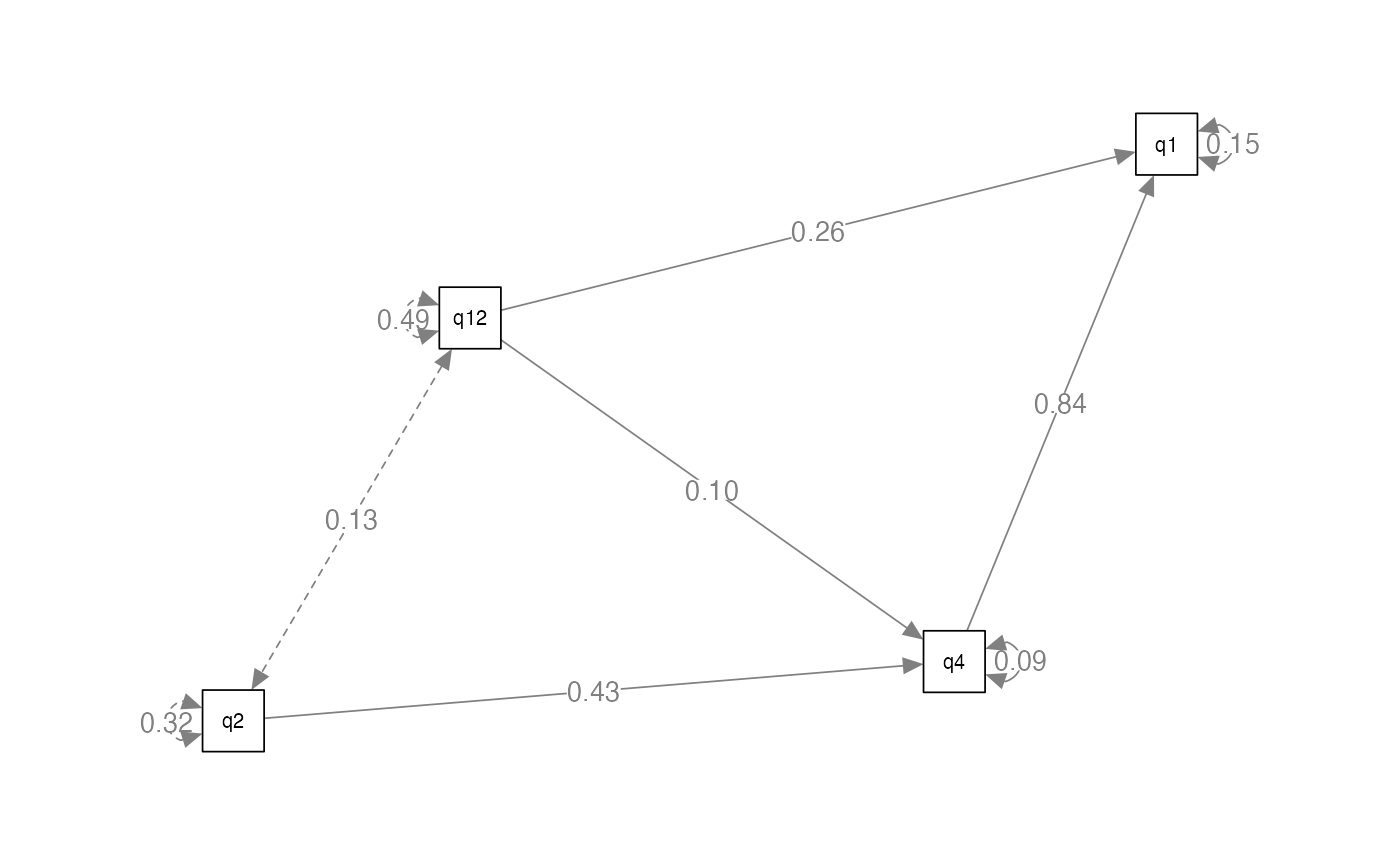
Another Example
- You don’t actually have to have the data to run an analysis
- If you have the correlation matrix, you can still run the analysis
- It’s always better to use the raw data if you have it!
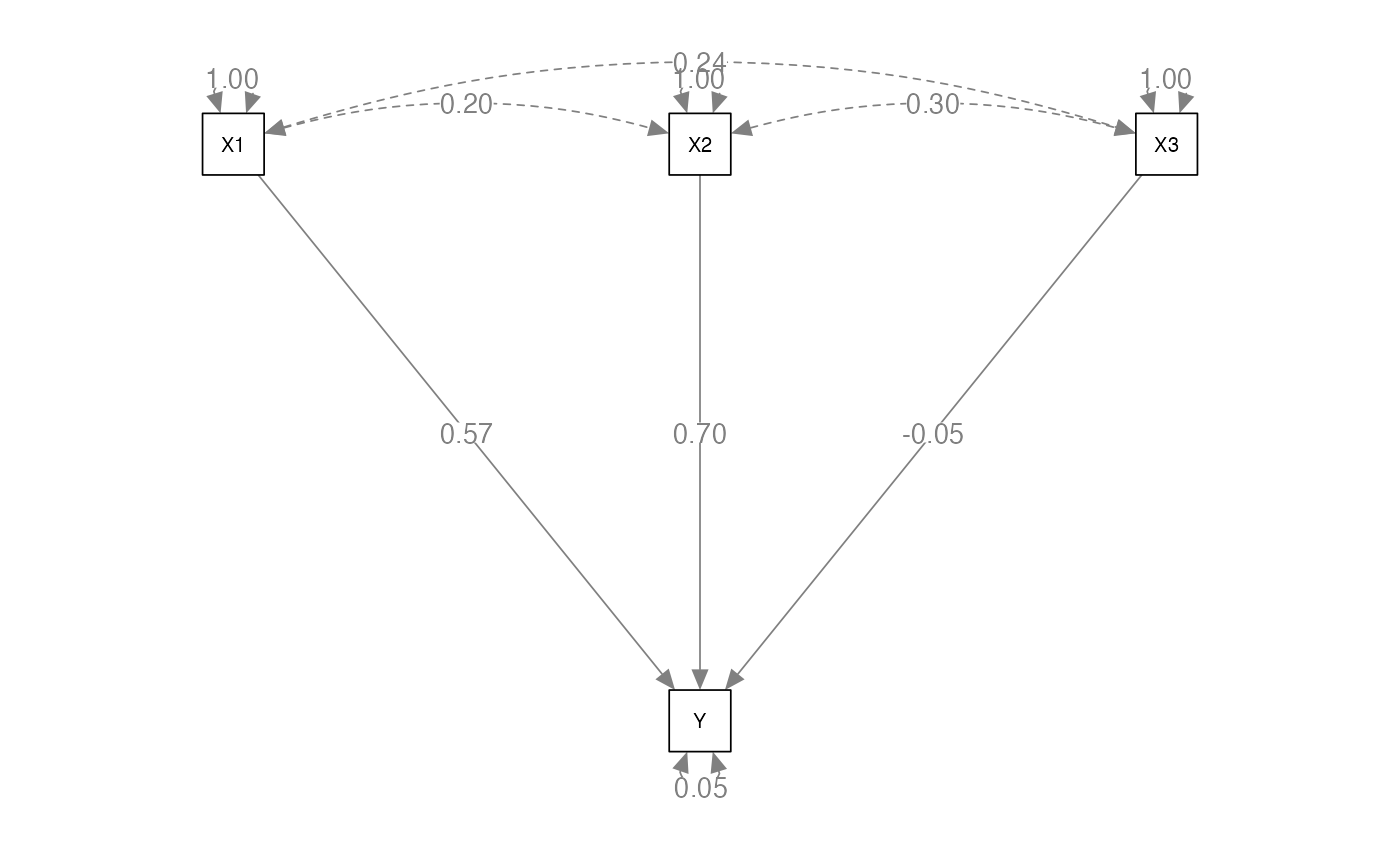
regression.cor <- lav_matrix_lower2full(c(1.00,
0.20,1.00,
0.24,0.30,1.00,
0.70,0.80,0.30,1.00))
# name the variables in the matrix
colnames(regression.cor) <-
rownames(regression.cor) <-
c("X1", "X2", "X3", "Y") Build the Model
- Let’s look at how we can label the paths
regression.model <- '
# structural model for Y
Y ~ a*X1 + b*X2 + c*X3
# label the residual variance of Y
Y ~~ z*Y
'Analyze the Model
regression.fit <- sem(model = regression.model,
sample.cov = regression.cor, # instead of data
sample.nobs = 1000) # number of data pointsView the Summary
summary(regression.fit,
standardized = TRUE,
fit.measures = TRUE,
rsquare = TRUE)
#> lavaan 0.6-19 ended normally after 1 iteration
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 4
#>
#> Number of observations 1000
#>
#> Model Test User Model:
#>
#> Test statistic 0.000
#> Degrees of freedom 0
#>
#> Model Test Baseline Model:
#>
#> Test statistic 2913.081
#> Degrees of freedom 3
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 1.000
#> Tucker-Lewis Index (TLI) 1.000
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) 38.102
#> Loglikelihood unrestricted model (H1) 38.102
#>
#> Akaike (AIC) -68.205
#> Bayesian (BIC) -48.574
#> Sample-size adjusted Bayesian (SABIC) -61.278
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.000
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.000
#> P-value H_0: RMSEA <= 0.050 NA
#> P-value H_0: RMSEA >= 0.080 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> Y ~
#> X1 (a) 0.571 0.008 74.539 0.000 0.571 0.571
#> X2 (b) 0.700 0.008 89.724 0.000 0.700 0.700
#> X3 (c) -0.047 0.008 -5.980 0.000 -0.047 -0.047
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .Y (z) 0.054 0.002 22.361 0.000 0.054 0.054
#>
#> R-Square:
#> Estimate
#> Y 0.946Mediation Models
- Mediation models are regression models that imply that two variables are related (X and Y) until you add the mediator variable (M), which diminishes the relationship between X and Y
- You can model these with regression or a structural layout
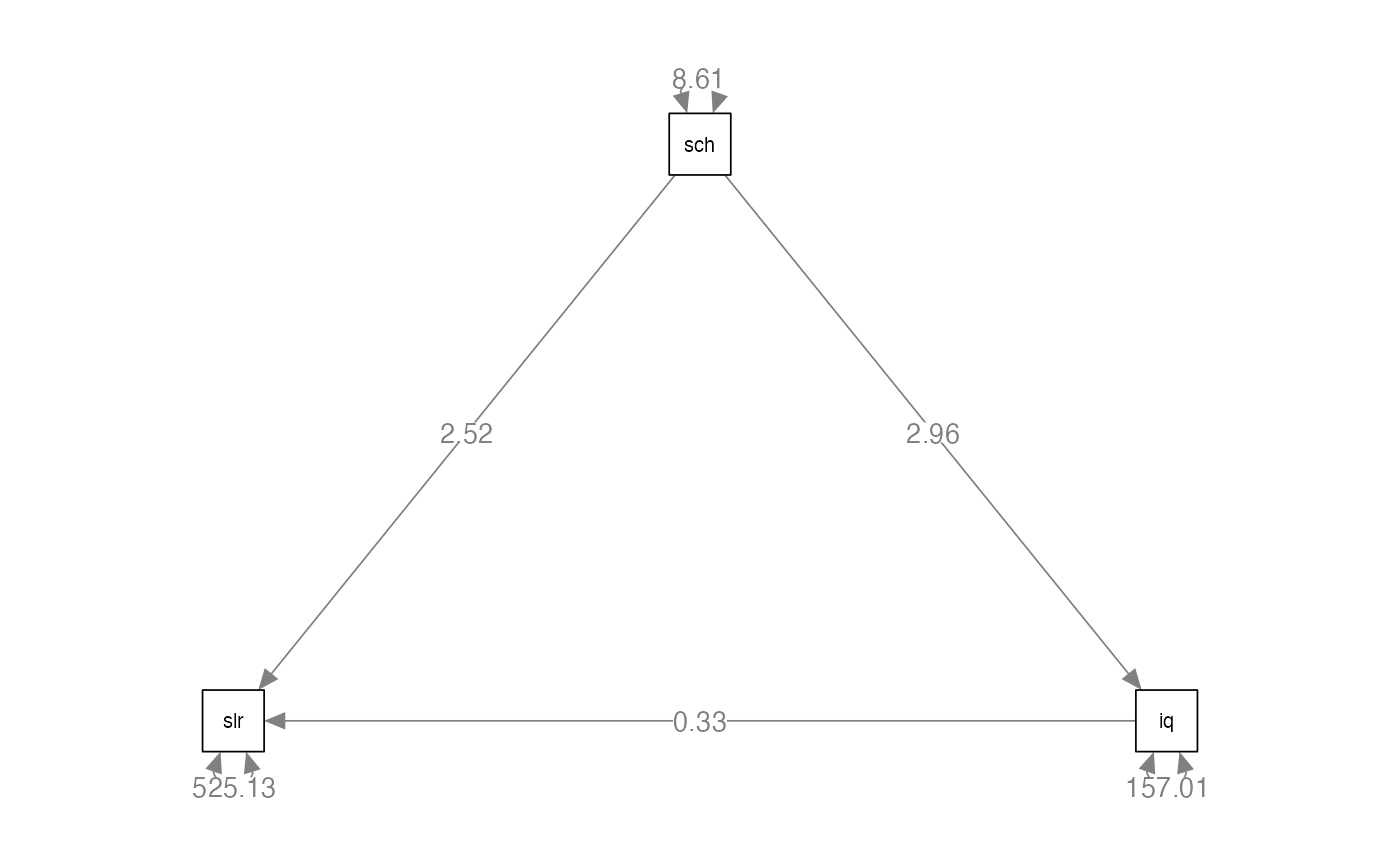
beaujean.cov <- lav_matrix_lower2full(c(648.07,
30.05, 8.64,
140.18, 25.57, 233.21))
colnames(beaujean.cov) <-
rownames(beaujean.cov) <-
c("salary", "school", "iq")Build the Model
beaujean.model <- '
salary ~ a*school + c*iq
iq ~ b*school # this is reversed in first printing of the book
ind:= b*c # this is the mediation part
'Analyze the Model
beaujean.fit <- sem(model = beaujean.model,
sample.cov = beaujean.cov,
sample.nobs = 300)View the Summary
summary(beaujean.fit,
standardized = TRUE,
fit.measures = TRUE,
rsquare = TRUE)
#> lavaan 0.6-19 ended normally after 1 iteration
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 5
#>
#> Number of observations 300
#>
#> Model Test User Model:
#>
#> Test statistic 0.000
#> Degrees of freedom 0
#>
#> Model Test Baseline Model:
#>
#> Test statistic 179.791
#> Degrees of freedom 3
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 1.000
#> Tucker-Lewis Index (TLI) 1.000
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -2549.357
#> Loglikelihood unrestricted model (H1) -2549.357
#>
#> Akaike (AIC) 5108.713
#> Bayesian (BIC) 5127.232
#> Sample-size adjusted Bayesian (SABIC) 5111.375
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.000
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.000
#> P-value H_0: RMSEA <= 0.050 NA
#> P-value H_0: RMSEA >= 0.080 NA
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.000
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> salary ~
#> school (a) 2.515 0.549 4.585 0.000 2.515 0.290
#> iq (c) 0.325 0.106 3.081 0.002 0.325 0.195
#> iq ~
#> school (b) 2.959 0.247 12.005 0.000 2.959 0.570
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .salary 525.128 42.877 12.247 0.000 525.128 0.813
#> .iq 157.011 12.820 12.247 0.000 157.011 0.676
#>
#> R-Square:
#> Estimate
#> salary 0.187
#> iq 0.324
#>
#> Defined Parameters:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> ind 0.963 0.323 2.984 0.003 0.963 0.111Fit Indices
- Now that we’ve done a few examples, how can we know if our model is representative of the data?
- We can use fit indices!
- There are a lot of them
- They do not imply that you are perfectly right
- They have guidelines but they are not perfect either
- People misuse them.
Limitations
- Fit statistics indicate an overall/average model fit
- That means there can be bad sections, but the overall fit is good
- No one magical number/summary
- They do not tell you where a misspecification occurs
- Do not tell you the predictive value of a model
- Do not tell you if it’s theoretically meaningful
“Rules”
- Everyone cites Hu and Bentler (1999) for the golden standards.
- Practically, these rules represent the same problems as rules about p values and effect sizes
- Generally the rules fall into two categories:
- Goodness of fit statistics: want values close to 1
- Excellent > .95, Good > .90
- Badness of fit statistics: want values close to 0
- Excellent < .06, Good < .08, Acceptable < .10
- Goodness of fit statistics: want values close to 1
Model Test Statistic
- Examines if the reproduced correlation matrix matches the sample correlation matrix
- Sometimes called “badness of fit”
- You want this value to be small as it measures the “mismatch” or error
Model Test Statistic
- Traditional null hypothesis testing reject-support context
- Generally, we reject the null to show that it’s unlikely, and therefore, the research hypothesis is likely
- However, here we want to support that the model represents the data, which is predicting the null
- So, it’s an odd use of to “support” the null
Model Test Statistic
-
- FML is the minimum fit function in maximum likelihood estimation
- p values are based on the df for your model and the chi-square distribution
- You want this value to be non-significant (minus the issues of predicting the null)
- However, this non-significance is problematic!
Model Test Statistic
- Chi-square is biased by:
- Multivariate non-normality
- Correlation size - large correlations can be hard to estimate
- Unique variance
- Sample size this is the big one
Model Test Statistic
- Everyone reports chi-square, but people tend to ignore significant
values
- One suggestion was normed chi-square - however, it is known to be just as problematic
- Since
is biased, you could use a robust estimator to correct for bias
- Satorra-Bentler
- Yuan-Bentler
Alternative Fit Indices
- There are three types of models:
- Your Model
- Independence - a model assuming no relationships between the variables (i.e., parameters are not significant)
- Saturated - a model assuming all parameters exist (i.e., df =
0).
- We can use a ratio of your model compared to these other possible models as a way to determine fit.
- These are less black and white, until we apply the “rules”
- It’s easy to “cherry-pick” the best indices to support your model
Alternative Fit Indices
- Absolute Fit Indices
- Incremental Fit Indices
- Parsimony-adjusted Indices
- Predictive Fit Indices
Absolute fit indices
- Proportion of the covariance matrix explained by the model
- You can think about these as sort of
- Want these values be high
- Goodness of Fit Index (GFI), Adjusted Goodness of Fit Index (AGFI),
Parsimonious Goodness of Fit Index (PGFI)
- Where residual is the variance not explained by the model, total is the total amount of variance
- Research indicates these are positively biased, and they are not recommended for use
Absolute Fit Indices
- Standardized root mean residual (SRMR)
- Want small values, as it’s a badness of fit index
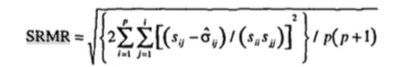
Incremental Fit Indices
- Also known as comparative fit indices
- Values are to 0 to 1, want high values
- Your model compared to the improvement over the independence model (no relationships)
- Comparative Fit Index (CFI)
- Normed Fit Index (NFI)
- A variation of the CFI, as it was said to underestimate for small samples
Incremental Fit Indices
- Incremental Fit Index (IFI)
- Also known as Bollen’s Non-normed fit index
- Modified NFI that decreases emphasis on sample size
- Relative Fit Index (RFI)
- Also known as Bollen’s Normed Fit Index
Incremental Fit Indices
- Tucker Lewis Index (TLI)
- Also known as the Bentler-Bonet Non-Normed Fit Index
- A popular fit index
Parsimony Adjusted Indices
- These include penalties for model complexity because added paths often result in better fit (overfitting)
- These will have smaller values for simpler models
- Root mean squared error of approximation (RMSEA)
- The most popular index
- Report the confidence interval
Predictive Fit Indices
- Estimate model fit in a hypothetical replication of the study with the same sample size randomly drawn from the population
- Most often used for model comparisons, rather than “fit”
- Often also considered parsimony adjusted indices
Model Comparisons
- Let’s say you want to adjust your model
- You can compare the adjusted model to the original model to determine if the adjustment is better
- Let’s say you want to compare two different models
- You can compare their fits to see which is better
Model Comparisons
- Nested models
- If you can create one model from another by the addition or subtraction of parameters, then it is nested
- Model A is said to be nested within Model B, if Model B is a more complicated version of Model A
- For example, a one-factor model is nested within a two-factor as a one-factor model can be viewed as a two-factor model in which the correlation between factors is perfect
Model Comparisons Nested Models
- Chi-square difference test - is this difference in models significant?
- If yes, you say the model with the lower chi-square is better
- If no, you say they are the same and go with the simpler model
chi_difference <- 12.6 - 4.3
df_difference <- 14 - 12
pchisq(chi_difference, df_difference, lower.tail = F)
#> [1] 0.01576442Model Comparisons Nested Models
- CFI difference test
- Subtract CFI model 1 - CFI model 2
- If the change is more than .01, then the models are considered different
- This version is not biased by sample size issues with chi-square
Model Comparisons Nested Models
- So how can I tell what to change?
- Just change one thing at a time!
- Modification indices
- Tell you what the chi-square change would be if you added the path suggested
- Based on (Lagrange Multiplier)
- Anything with > 3.84 is p < .05
Model Comparisons Nested Models
- Instead of doing the math, you can use the
anova()function anova(model1.fit, model2.fit)
Model Comparisons Non-Nested Models
- Akaike Information Criterion (AIC)
- Bayesian Information Criterion (BIC)
- Sample size Adjusted Information Criterion (SABIC)
- All of these penalize you for having more complex models.
- If all other things are equal, it is biased to the simpler model.
- To compare, pick the model with the lower value (always lower, even when negative)
Model Comparisons Non-Nested Models
- Expected Cross Validation Index (ECVI)
- t is the number of parameters estimated
- p is the number of squares (measured variables)
- Same rules, pick the smallest value
So what to report?
- We will use RMSEA, SRMR, CFI
- Generally, people also report chi-square and df
- Determine the type of model change to use the right model comparison statistic
Path Comparison Example
compare.data <- lav_matrix_lower2full(c(1.00,
.53, 1.00,
.15, .18, 1.00,
.52, .29, -.05, 1.00,
.30, .34, .23, .09, 1.00))
colnames(compare.data) <-
rownames(compare.data) <-
c("morale", "illness", "neuro", "relationship", "SES") Build a Model
#model 1
compare.model1 = '
illness ~ morale
relationship ~ morale
morale ~ SES + neuro
'
#model 2
compare.model2 = '
SES ~ illness + neuro
morale ~ SES + illness
relationship ~ morale + neuro
'Analyze the Model
compare.model1.fit <- sem(compare.model1,
sample.cov = compare.data,
sample.nobs = 469)
summary(compare.model1.fit,
standardized = TRUE,
fit.measures = TRUE,
rsquare = TRUE)
#> lavaan 0.6-19 ended normally after 5 iterations
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 8
#>
#> Number of observations 469
#>
#> Model Test User Model:
#>
#> Test statistic 40.303
#> Degrees of freedom 4
#> P-value (Chi-square) 0.000
#>
#> Model Test Baseline Model:
#>
#> Test statistic 390.816
#> Degrees of freedom 9
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 0.905
#> Tucker-Lewis Index (TLI) 0.786
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -1819.689
#> Loglikelihood unrestricted model (H1) -1799.537
#>
#> Akaike (AIC) 3655.377
#> Bayesian (BIC) 3688.582
#> Sample-size adjusted Bayesian (SABIC) 3663.192
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.139
#> 90 Percent confidence interval - lower 0.102
#> 90 Percent confidence interval - upper 0.180
#> P-value H_0: RMSEA <= 0.050 0.000
#> P-value H_0: RMSEA >= 0.080 0.995
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.065
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> illness ~
#> morale 0.530 0.039 13.535 0.000 0.530 0.530
#> relationship ~
#> morale 0.520 0.039 13.184 0.000 0.520 0.520
#> morale ~
#> SES 0.280 0.045 6.217 0.000 0.280 0.280
#> neuro 0.086 0.045 1.897 0.058 0.086 0.086
#>
#> Covariances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .illness ~~
#> .relationship 0.014 0.033 0.430 0.667 0.014 0.020
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .illness 0.718 0.047 15.313 0.000 0.718 0.719
#> .relationship 0.728 0.048 15.313 0.000 0.728 0.730
#> .morale 0.901 0.059 15.313 0.000 0.901 0.903
#>
#> R-Square:
#> Estimate
#> illness 0.281
#> relationship 0.270
#> morale 0.097Analyze the Model
compare.model2.fit <- sem(compare.model2,
sample.cov = compare.data,
sample.nobs = 469)
summary(compare.model2.fit,
standardized = TRUE,
fit.measures = TRUE,
rsquare = TRUE)
#> lavaan 0.6-19 ended normally after 1 iteration
#>
#> Estimator ML
#> Optimization method NLMINB
#> Number of model parameters 9
#>
#> Number of observations 469
#>
#> Model Test User Model:
#>
#> Test statistic 3.245
#> Degrees of freedom 3
#> P-value (Chi-square) 0.355
#>
#> Model Test Baseline Model:
#>
#> Test statistic 400.859
#> Degrees of freedom 9
#> P-value 0.000
#>
#> User Model versus Baseline Model:
#>
#> Comparative Fit Index (CFI) 0.999
#> Tucker-Lewis Index (TLI) 0.998
#>
#> Loglikelihood and Information Criteria:
#>
#> Loglikelihood user model (H0) -1796.138
#> Loglikelihood unrestricted model (H1) -1794.516
#>
#> Akaike (AIC) 3610.276
#> Bayesian (BIC) 3647.632
#> Sample-size adjusted Bayesian (SABIC) 3619.068
#>
#> Root Mean Square Error of Approximation:
#>
#> RMSEA 0.013
#> 90 Percent confidence interval - lower 0.000
#> 90 Percent confidence interval - upper 0.080
#> P-value H_0: RMSEA <= 0.050 0.742
#> P-value H_0: RMSEA >= 0.080 0.050
#>
#> Standardized Root Mean Square Residual:
#>
#> SRMR 0.016
#>
#> Parameter Estimates:
#>
#> Standard errors Standard
#> Information Expected
#> Information saturated (h1) model Structured
#>
#> Regressions:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> SES ~
#> illness 0.309 0.043 7.110 0.000 0.309 0.309
#> neuro 0.174 0.043 4.019 0.000 0.174 0.174
#> morale ~
#> SES 0.135 0.041 3.291 0.001 0.135 0.135
#> illness 0.484 0.041 11.756 0.000 0.484 0.484
#> relationship ~
#> morale 0.540 0.039 13.745 0.000 0.540 0.538
#> neuro -0.131 0.039 -3.335 0.001 -0.131 -0.131
#>
#> Variances:
#> Estimate Std.Err z-value P(>|z|) Std.lv Std.all
#> .SES 0.853 0.056 15.313 0.000 0.853 0.855
#> .morale 0.701 0.046 15.313 0.000 0.701 0.703
#> .relationship 0.711 0.046 15.313 0.000 0.711 0.710
#>
#> R-Square:
#> Estimate
#> SES 0.145
#> morale 0.297
#> relationship 0.290View the Model
semPaths(compare.model1.fit,
whatLabels="par",
edge.label.cex = 1,
layout="spring")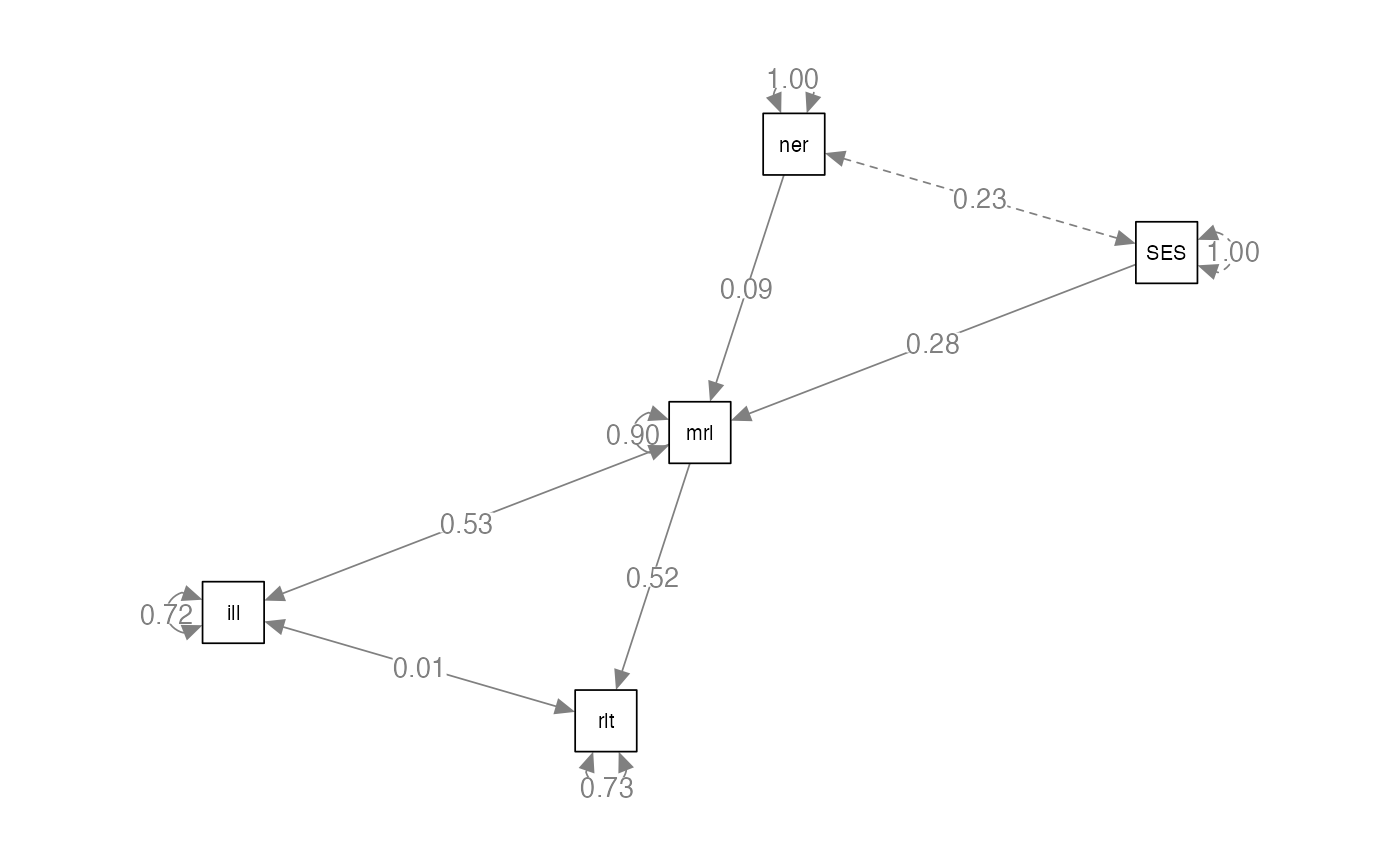
semPaths(compare.model2.fit,
whatLabels="par",
edge.label.cex = 1,
layout="spring")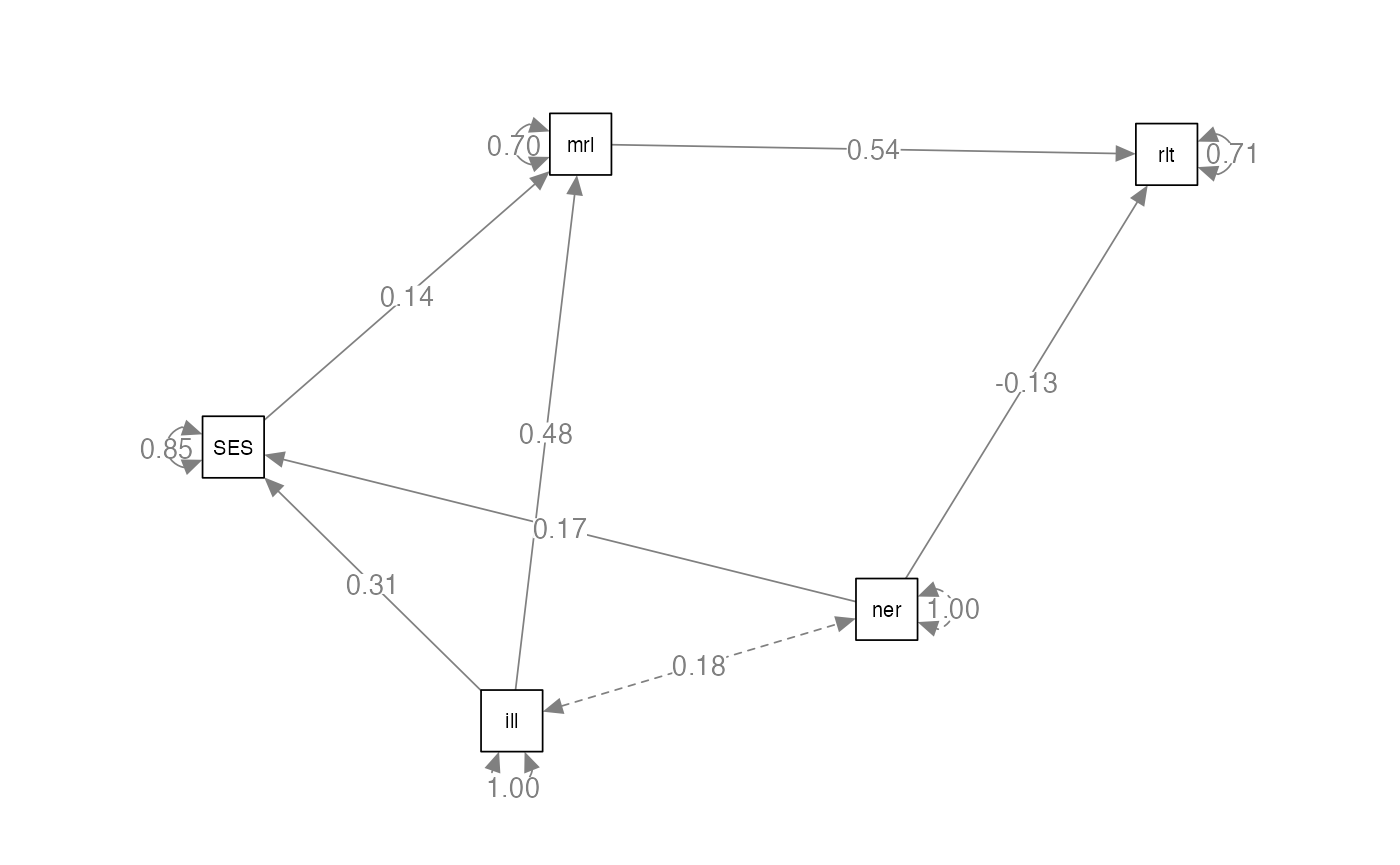
Compare the Models
anova(compare.model1.fit, compare.model2.fit)
#>
#> Chi-Squared Difference Test
#>
#> Df AIC BIC Chisq Chisq diff RMSEA Df diff
#> compare.model2.fit 3 3610.3 3647.6 3.2454
#> compare.model1.fit 4 3655.4 3688.6 40.3031 37.058 0.27728 1
#> Pr(>Chisq)
#> compare.model2.fit
#> compare.model1.fit 1.147e-09 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
fitmeasures(compare.model1.fit, c("aic", "ecvi"))
#> aic ecvi
#> 3655.377 0.120
fitmeasures(compare.model2.fit, c("aic", "ecvi"))
#> aic ecvi
#> 3610.276 0.045